Our Location
Sector 14, Dwarka, New Delhi
A clinically validated NGS panel with optimized bioinformatics for analyzing germline mutations associated with hereditary cancers – including Lynch syndrome.
Complete hereditary cancer analysis at your fingertips. Streamline your testing from sample to data analysis.

Cancer type |
Recommended genes for screening included in GALEAS HereditaryPlus |
|
| Breast | ATM, BARD1, BRCA1, BRCA2, CDH1, CHEK2, NBN, NF1, PALB2, PTEN, STK11, TP53 | |
| Colon | APC, AXIN2, BMPR1A, CHEK2, EPCAM, GREM1, MLH1, MSH2, MSH6, PMS2, MSH3, MUTYH, NTLH1, POLD1, POLE, PTEN, RNF43, SMAD4, STK11, TP53 | |
| Renal | BAP1, FH, FLCN, MET, SDHB, VHL | |
| Ovarian | ATM, BARD1, BRCA1, BRCA2, CDH1, CHEK2, NBN, NF1, PALB2, PTEN, SKT11, TP53, RAD51C, RAD51D | |
| Prostate | ATM, BRCA1, BRCA2, CHEK2, MLH1, MSH2, MSH6, PALB2 | |
| Gastric/GIST | CDH1, KIT, PDGFRA, SDHC, SDHD, SDHA | |
| Brain | APC, ATM, MLH1, MSH2, MSH6, PMS2, TP53 | |
| Sarcoma | EXT1, EXT2, MTAP, NF1, RECQL4, SQSTM1, TP53 | |
| Pediatric (Wilms Tumor) | CDKN1C, CTR9, REST, TRIM28, WT1 | |
With GALEAS HereditaryPlus, laboratories can trust in exceptional accuracy and reliability of variant detection across a wide range of alteration types. Our commitment to innovation and precision empowers clinicians to make informed decisions and provide personalized care to individuals at risk of hereditary cancer.
ID |
Gene |
HGVS coding |
HGVS protein |
Genomic position |
| 22 | BRCA1 | c.1175_1214del | p.Leu392fs*5 | chr17:43094317 |
| 23 | BRCA1 | c.1175_1214del | p.Leu392fs*5 | chr17:43094317 |
| 64 | MSH2 | c.942+3A>T | p.? | chr2:47414421 |
| 65 | PMS2 | c.736_741delinsTGTGTGTGAAG | p.(Pro246Cysfs*3) | chr7:5997389 |
| 66 | MLH1 | c.1946dupC | p.(Leu650Phefs*14) | chr3:37048561 |
| 67 | MSH2 | c.1213_1217dup | p.(Leu407Thrfs*7) | chr2:47429877 |
| 68 | MSH6 | c.3562_3563del | p.(Ser1188Tyrfs*5) | chr2:47805623 |
Table 2. SNV recall was shown to be 100% across a wide range of alteration types including small and large indels
Overcoming Challenges: CNVs, mosaics and pseudogenes

In a clinical study of 64 patients with orthogonal data, GALEAS HereditaryPlus delivered an analytical sensitivity of 98.3% and an analytical specificity of 99% for CNV analysis. This high level of sensitivity and specificity guarantees the identification of critical genetic alterations associated with hereditary cancer, empowering clinicians with actionable information for disease management.
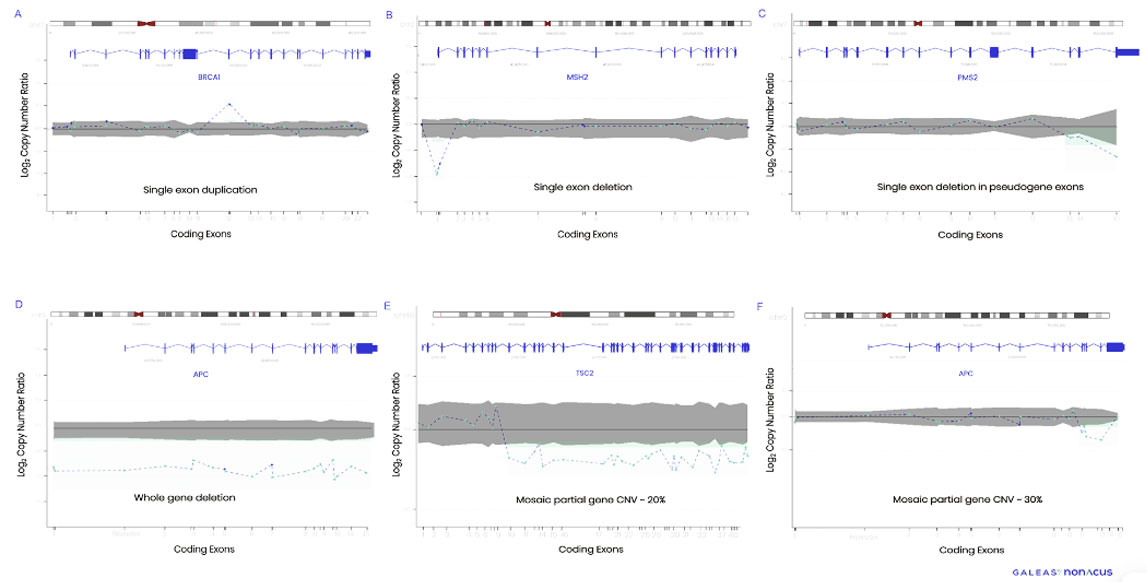
Figure 1. CNV profiles detected by GALEAS HereditaryPlus. A) BRCA1 single exon duplication, B) MSH2 single exon deletion, C) PMS2 multiple exon deletion in pseudogene D) APC whole gene deletion, E) TSC2 mosaic partial gene CNV -20%, F) APC mosaic partial gene CNV- 30%.
GALEAS HereditaryPlus eliminates the need for time-consuming and costly supplementary tests providing laboratories with a consolidated and efficient testing workflow for hereditary cancer.
Cloud-based GALEAS Analysis software delivers a comprehensive variant calling pipeline.Features of GALEAS HereditaryPlus:
Avoid unnecessary ancillary testing like MLPA or Sanger sequencing
Confidently call all variants including MSH2 c.942+3A>T variant a wide range of CNVs in genes such as APC, MHS2, BRCA1 and PMS2 with one NGS based workflow.
Validate and run a single workflow for all hereditary cancers.
Consolidate workflows for all hereditary cancers including breast, prostate, Lynch syndrome and Wilms tumor, reducing operational time and cost.
Streamline bioinformatics with GALEAS Analysis Software
Optimized for the GALEAS HereditaryPlus panel, our cloud-based bioinformatics pipelines deliver the accurate variant calling critical for reliable downstream interpretation.
Key quality indicator |
GALEAS HereditaryPlus |
Company I |
| Number of Genes | 146 | 113 |
| Capture Panel size (kb) | 809 | 403 |
| MB required for mean 100x coverage | 130 MB | 116.6 MB |
| Percentage coverage >30x | 99% | 96% |
| Percentage on bait | 71.2% | 37.0% |
| Percentage on or near bait | 81% | 61.51% |
| Percent duplication | 2.0% | 8.99% |
| SNV recall | 99.7% | 98.1% |
| Indel Recall | 100% | 97.2% |
gDNA samples
Wide range of sample types
Prepare samples
BeadXtract DNA extraction kits
Prepare libraries and enrich
GALEAS HereditaryPlus
Sequence
Illumina NGS Sequencing System
Call variants
GALEAS Analysis software
Interpret and report
Secondary software for interpretation and reporting
Parameters |
Specification |
| Enrichment method | Hybridization and capture |
| Number of genes | 146 |
| Capture Panel size | 809 kb |
| Sequencing platform | Illumina |
| Targets | Genes associated with hereditary cancer |
| Variant types | SNVs, CNVs and INDELs |
| Input DNA requirements* | 10ng-200ng |
| Sample type | gDNA from blood or saliva |
| Multiplexing guidance for sequencing | 500K reads per sample required to achieve 100x. This equates to 0.15Gb per sample |
Product Description |
Pack Size |
Catalogue Number |
| GALEAS HereditaryPlus | 16 samples | NGS_GAL_HCP_FR_16 |
| (with enzymatic fragmentation for gDNA) | 96 samples | NGS_GAL_HCP_FR_96_A/B/C/D* |
For any queries or detailed requirements, get in touch with our team of experts at Shiva Scientific.








Shiva Scientific
Copyright © 2024. All rights reserved.