Detect all sequence variants
If a genetic testing workflow is terminated when a first sequence variant is found, or a method is used where only sequence variant-specific detection is possible, there is a risk that additional sequence variants remain undetected.
A recent study shows that the Devyser Thalassemia NGS assay detects sequence variants that were not discovered with traditional Thalassemia testing. Devyser’s kit was used to analyse 125 samples previously characterised to carry pathogenic sequence variants in the globin genes using traditional genetic testing methods and workflows.
-
- All previously detected pathogenic SNVs, Indels and CNVs in the globin genes were confirmed using Devyser Thalassemia.
-
- In 15% of samples, additional pathogenic sequence variants were found. The majority of these were found in a different globin gene. These sequence variants had previously not been identified since the workflow had been terminated as soon as a pathogenic sequence variant was found in the first gene cluster investigated.
Genes and regions covered
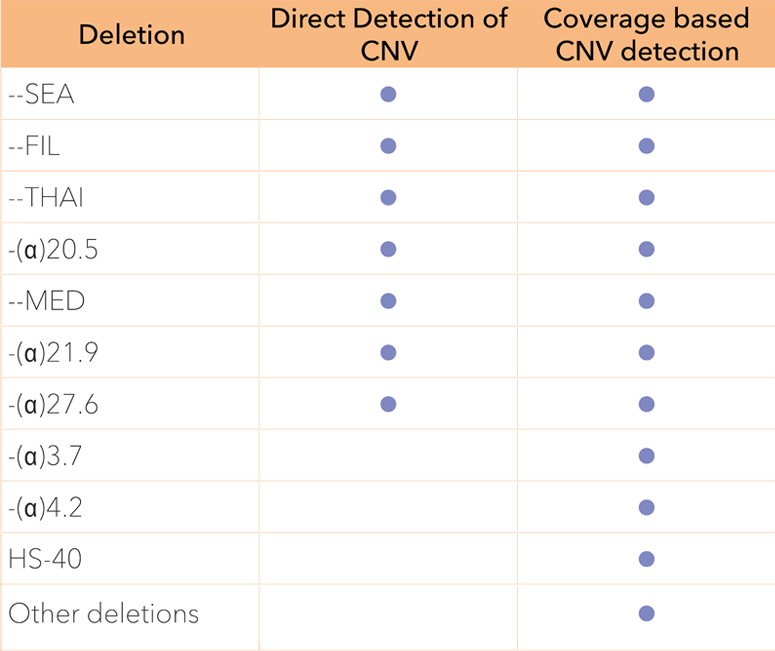
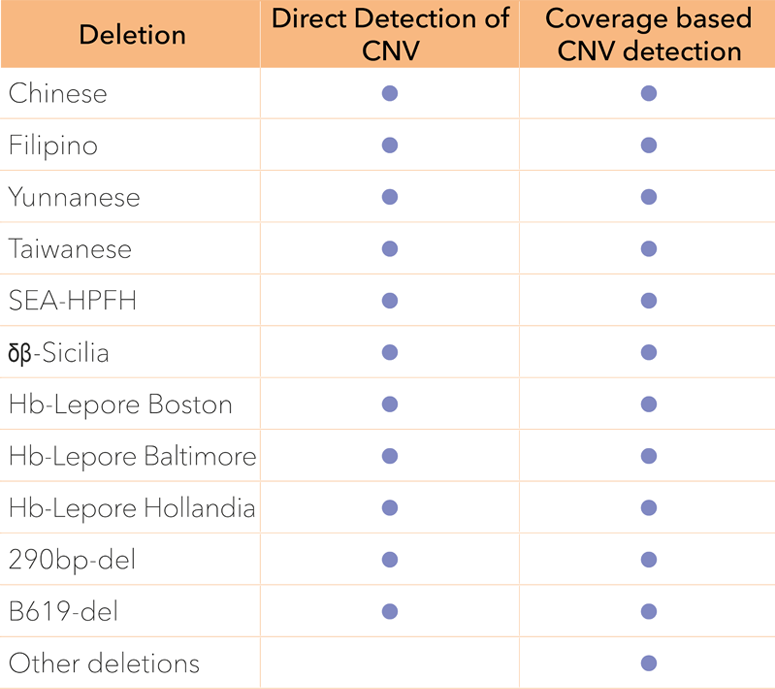
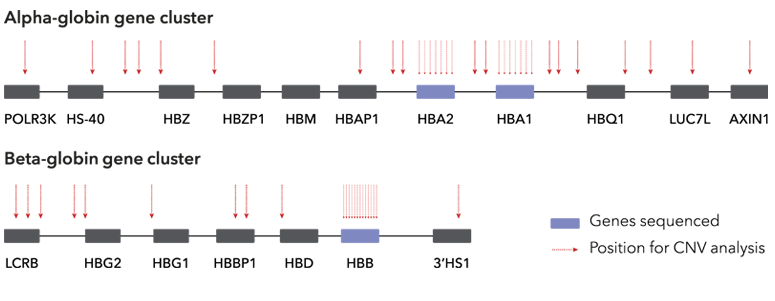
Large structural deletions are analysed over the entire alpha and beta globin gene clusters. The HBA1, HBA2 and HBB genes have an extra high coverage density to allow precise deletion mapping.
Genes colored in blue are fully sequenced to allow comprehensive detection of Indels and point sequence variants in the globin genes. Vertical arrows illustrate positions where the presence of CNVs (i.e. large structural deletions) are investigated.

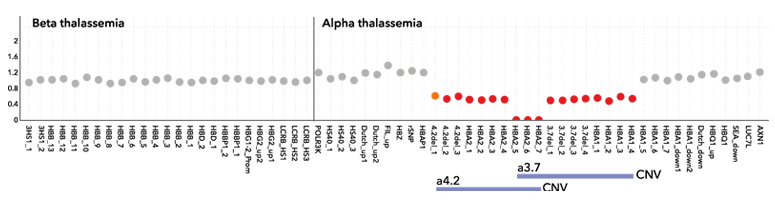
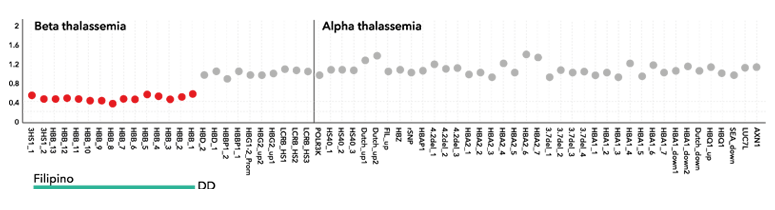 A sample with the Filipino deletion in the beta globin genes detected by direct detection of CNVs
A sample with the Filipino deletion in the beta globin genes detected by direct detection of CNVs